The origin of the language of life
19. Dezember 2014The genetic code is the universal language of life. It describes how information is encoded in the genetic material and is the same for all organisms from simple bacteria to animals to humans. However, the origin of the code remains enigmatic. Over the past two years, the research team of Bojan Žagrović at the Max F. Perutz Laboratories (MFPL) of the University of Vienna and the Medical University of Vienna has revealed several surprising clues that may help unlock this mystery. The results are described in a series of articles – the latest was published in December in Nucleic Acids Research.
All information about living organisms is stored in their genes. Genes, for example, encode our height, eye color or predisposition for different diseases. For the cell to be able to read the information contained in genes, each of them is first carbon-copied into a so-called messenger RNA (mRNA), whose content is then translated to make proteins, biology’s workhorses. While the information in mRNAs is encoded using a 4-letter alphabet of nucleobases, proteins are built using a 20-letter alphabet of amino acids. Scientists have deciphered the language used for translating mRNAs to proteins, the genetic code, already 50 years ago. Although we now know how to read the words made of nucleobases and understand which amino acids they stand for, the origin of this universal language of life remains mysterious.
Coding from binding
One idea, the so-called stereochemical hypothesis, suggests that the code may be a consequence of direct binding propensities of amino acids for the appropriate nucleobases. Simply put, the hypothesis proposes that symbols in the genetic code (nucleobases) may directly bind to the objects they stand for (amino acids). As such propensities appear to be quite weak, however, convincing experimental evidence in support of the hypothesis has been difficult to come by.
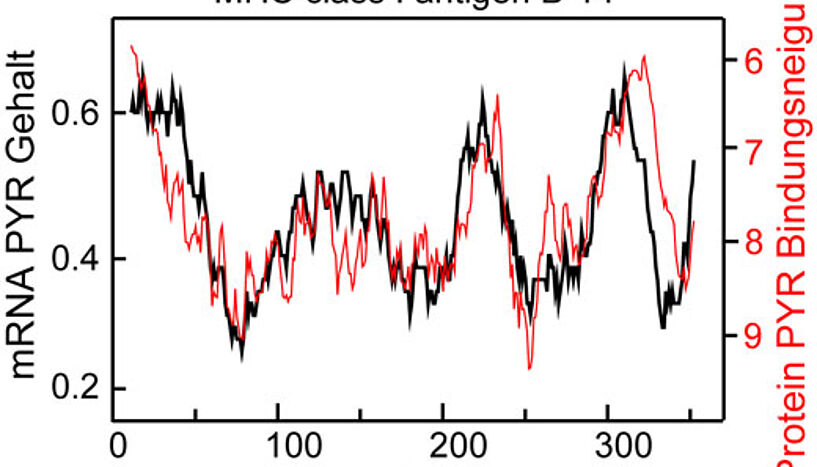
On the other hand, one might get amplified signals if one looks at complete mRNAs and the proteins they code for – a possibility that ERC Starting Grant awardee Bojan Žagrović and his coworkers at the Max F. Perutz Laboratories (MFPL) of the University of Vienna have been exploring over the past two years. The team has collected evidence that most mRNAs in modern organisms indeed exhibit signatures of potential complementary binding with the proteins they code for. The researchers studied this relationship for complete proteomes – the entire sets of proteins – of 15 different organisms, covering all three domains of life.
Complementary interactions
Using experimentally and computationally derived data, they found that the density profiles of different nucleobases in mRNAs closely resemble the profiles of amino-acid affinity for these same nucleobases in the proteins they encode. Moreover, they showed that the genetic code is highly optimized to maximize such matching. Importantly, this not only suggests that the genetic code could indeed have evolved as a consequence of direct binding interactions between mRNAs and proteins, but also implies that such interactions might still be relevant in present-day organisms. The latter would represent a still unknown layer of gene regulation.
Similarities to a cookie mold
“Imagine sending a group of friends a recipe for baking a cookie in the shape of a Christmas tree. If the look and shape of the tree were explained to them in writing, no tree would look exactly the same. But if one sent all of them a tree-shaped cookie mold, all trees would be identical: information about the shape of the cookie is faithfully encoded in the complementary shape of its mold. Our results suggest that the genetic code could have had a simple beginning whereby information about protein sequences was stored in the physiochemically complementary mRNA sequences. It is possible that our analysis has in fact revealed the vestiges of such a bygone era in present-day biomolecules”, explains Bojan Žagrović.
Do echoes of primordial events still resonate today?
While the majority of mRNA nucleobases were shown to have direct preference for amino acids they tend to encode, only one, adenine, exhibits the opposite behavior. This may indicate that adenine entered the genetic code during the second stage of its development in order to modulate and weaken binding. The scientists are now looking for experimental evidence to support these claims, but are also further analyzing the potential role that direct mRNA-protein interactions may play in present-day biology. “The most obvious process during which mRNAs and their cognate proteins are present in close vicinity is translation. The newly synthesized protein could, for example, suppress the translation of its own mRNA by competing for binding with the translation machinery”, says Bojan Žagrović. “We continue to research if our results are indeed relevant for such regulatory processes in today’s cells. It would be very interesting if the echoes of primordial events still resonated within the fabric of the biology of today”, concludes Žagrović.
References
1. Anton A Polyansky, Mario Hlevnjak and Bojan Zagrovic: Proteome-wide analysis reveals clues of complementary interactions between mRNAs and their cognate proteins as the physicochemical foundation of the genetic code. RNA Biology. August 2013. DOI: http://dx.doi.org/10.4161/rna.25977
2. Matea Hajnic, Juan Iregui Osorio and Bojan Zagrovic: Computational analysis of amino acids and their sidechain analogs in crowded solutions of RNA nucleobases with implications for the mRNA–protein complementarity hypothesis. Nucleic Acids Research. December 2014. DOI: http://dx.doi.org/10.1093/nar/gku1035
Scientific contact
Ass.-Prof. Dr. Bojan Žagrović
Max F. Perutz Laboratories
Department for Structural and Computational Biology
University of Vienna
T +43-1-4277- 522 71
bojan.zagrovic(at)univie.ac.at
Press contact
Dr. Lilly Sommer
Max F. Perutz Laboratories
Communications
T +43-1-4277-240 14
lilly.sommer(at)mfpl.ac.at
Wissenschaftlicher Kontakt
Dr. Bojan Zagrovic
Max F. Perutz LaboratoriesUniversität Wien
1030 - Wien, Dr. Bohr-Gasse 9
+43-1-4277-522 71
bojan.zagrovic@univie.ac.at
Rückfragehinweis
Dr. Lilly Sommer
Max F. Perutz Laboratories, CommunicationsUniversität Wien
1030 - Wien, Dr.-Bohr-Gasse 9
+43-1-4277-240 14
lilly.sommer@univie.ac.at